Binary Autoregressive Network Modeling of Comorbidity Networks from Electronic Health Records
Xi (Rossi) LUO
Health Science Center
School of Public Health
Dept of Biostatistics
and Data Science

December 15, 2020
Funding: NIH R01EB022911 and UT Health Start-up Fund
Slides viewable on web:
bit.ly /ehrnet20
or
BigComplexData.com
Co-Authors

Gen Zhu
UT Health, BADS

Hulin Wu
UT Health, BADS
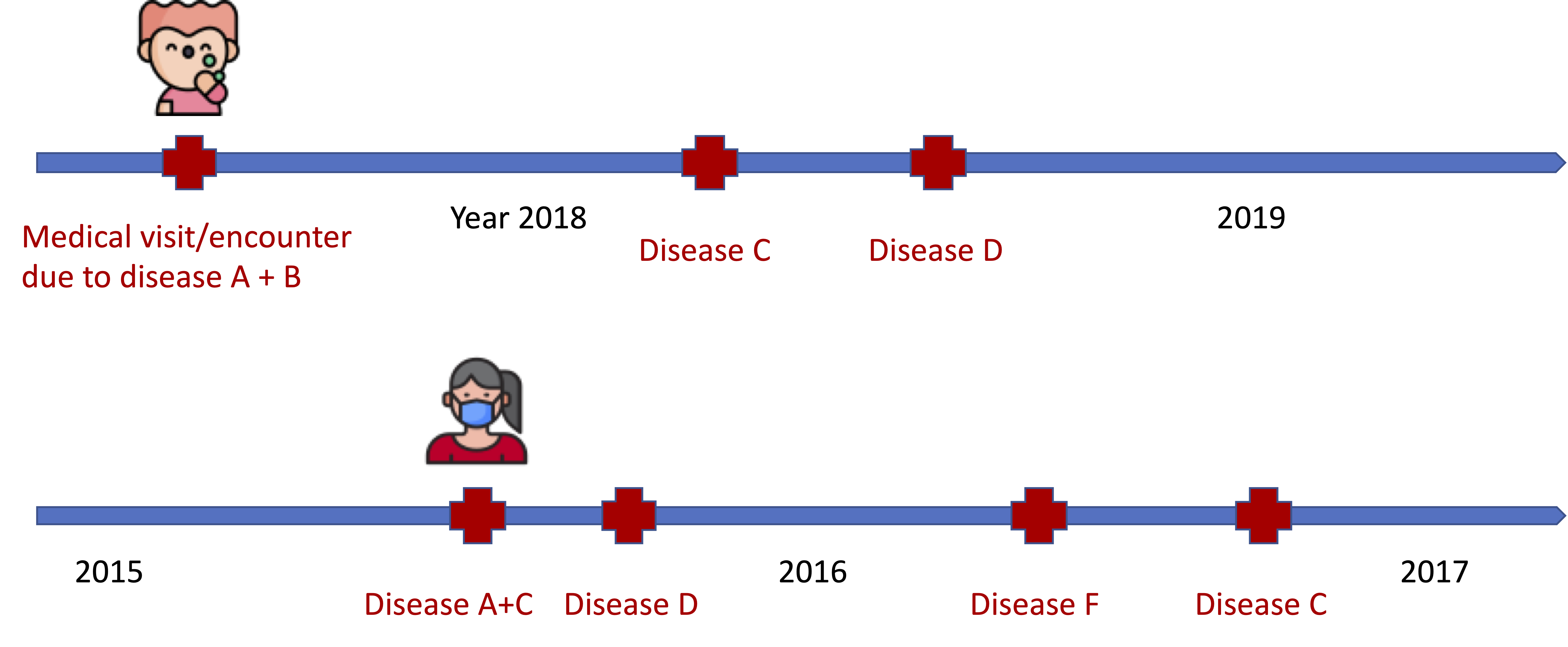
EHR Data: Medical Encounter/Diagnosis
Challenges
- Many many unique diagnosis codes (~100K)
- Large but heterogeneous samples (~10K to ~10M)
- In a nutshell, time series of events from a huge number of types
- Many other associated data types (lab, prescription)
Method
Existing Methods for Inferring Comorbidity Networks
- Most existing methods are pair-wise Fotouhi et al. 2018
- $w_{ij}$ be freq of disease $i$ happens prior to disease $j$
- Define link weights: $$s_x^{o} = \sum_y w_{xy}, \quad s_x^{i} = \sum_y w_{xy}, \quad, s = \sum_{xy} w_{xy}, $$
- $\phi$-correlation and OER: $$ \phi_{ij} = \frac{w_{ij} s - s_i^{o} s_j^{i}}{\sqrt{s_i^{o} s_j^{i} (s - s_i^{o}) (s - s_i^{i}) }}, \quad OER_{ij} = \frac{w_{ij}s}{s_j^{i} s_i^{o}} $$
- Univariate logistic regression Aguado et al. 2020
- First talk in this session by
Dr Maroufy and colleagues
Limitations
- Pair-wise associations fail to adjust other intermediate diseases developed in-between
- Multiple testing issues due to a large number of diseases $O(p^2)$
- Partially account for the temporal order
- Disease A, B, C may happen in a specific temporal order
Model
- We use ICD-9 codes for diagnoses
- $y_{ijk} = 1$ if patient $i$ has diagnosis code $k$ at encounter $j$, vector $Y_{ij}$ for all diagnosis codes
- Also known as one-hot encoding
- Binary autoregressive model $$ P(y_{ijk} = 1 | Y_{i,j-1}) = (1 + \exp(-Y_{i,j-1}^T \beta_k ) )^{-1} $$
- Inspired by Granger/vector autoregressive models for continuous variables
- $\beta_k$ denotes how each past diesase predicts future diagnosis $k$
Conditional Likelihood
- Full likelihood is challenging to compute
- Propose to optmize the penalized log-likelihood: $$\min_{\beta_k} \sum_{ij} \ell(y_{ijk} | \beta_k ) + \lambda \| \beta_k \|_1 $$
- Similar to Ising graphical models for binary data
without temporal ordering Ravikumar et al, 10; van de Geer et al, 14 - Implementation: LASSO penalized logistic regression
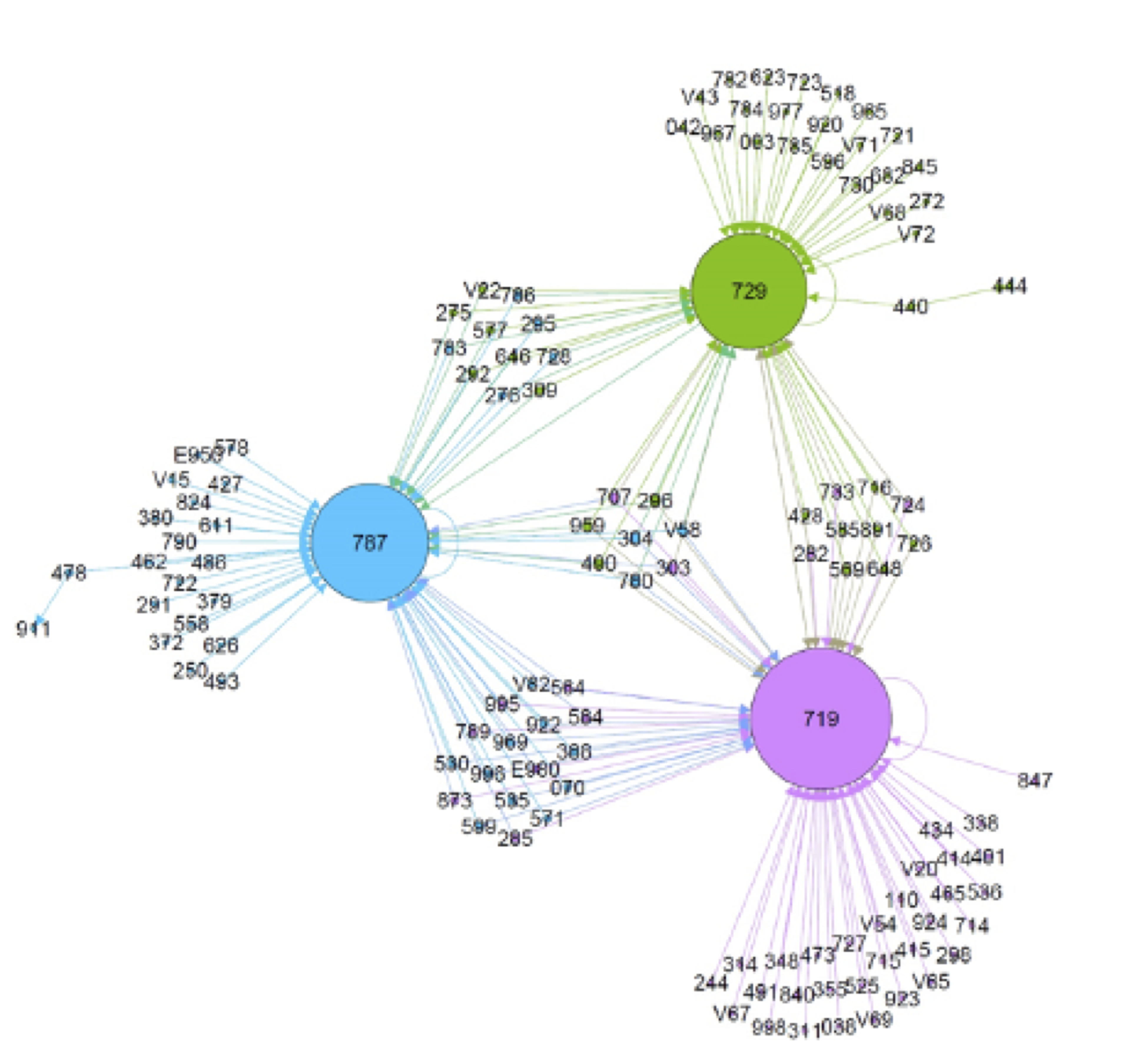
Real Data
Cerner's EHR
- Purchased EHR data by UT Health, Center for Big Data in Health Sciences, Director Dr. Hulin Wu
- Huge dataset: >60M paitients, ~1 billion diagnoses
Small dataset of patients with drug overdose diagnosis- 640 diseases, 11481 patients
- Goal: find network of diseases prior or after drug overdose
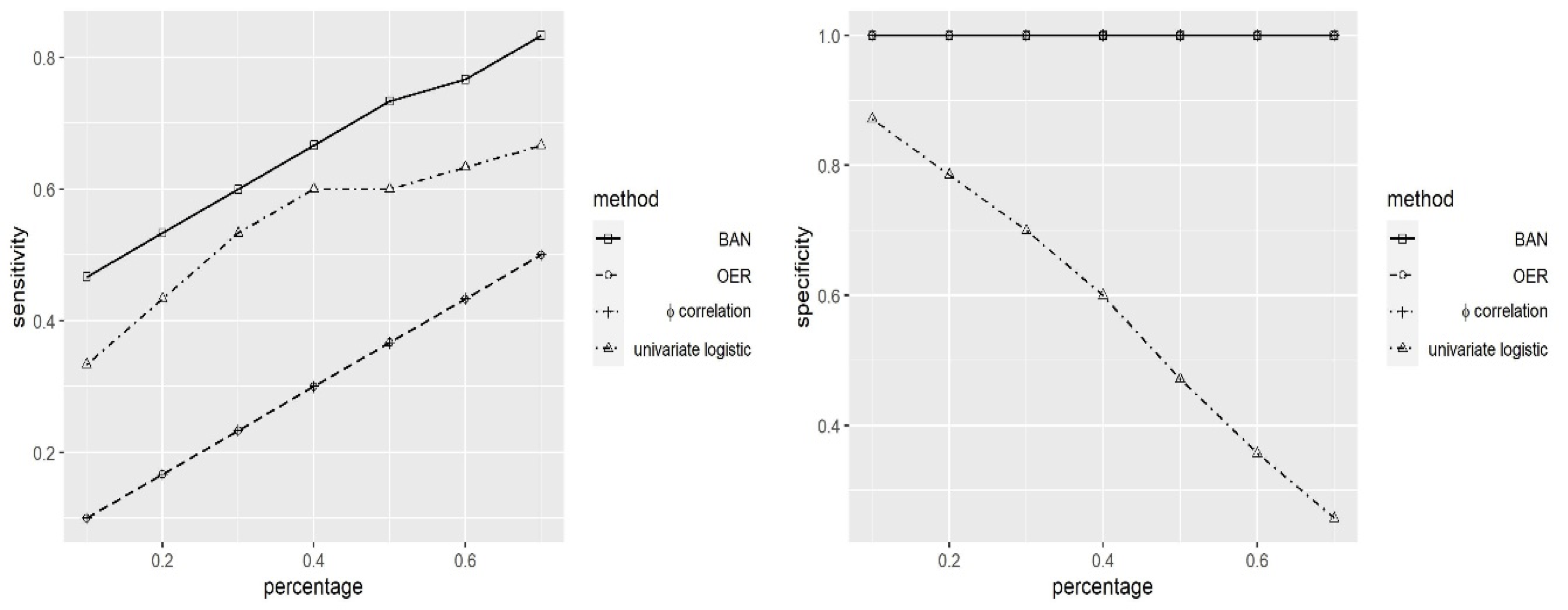
Simulations
Comparision with Other Methods
Discussion
- Model inspired by real-world EHR data
- Recovered directional disease networks
- Method: Granger causality + Ising models + ML
- high dimensionality, sparsity and temporality
- Many future directions:
- Bottle neck: managing and extracting data
- Lots of opportunities for theory and method
Thank you!
Comments? Questions?
BigComplexData.com